MicrobioKGs
Knowledge Graph applications in Microbiology
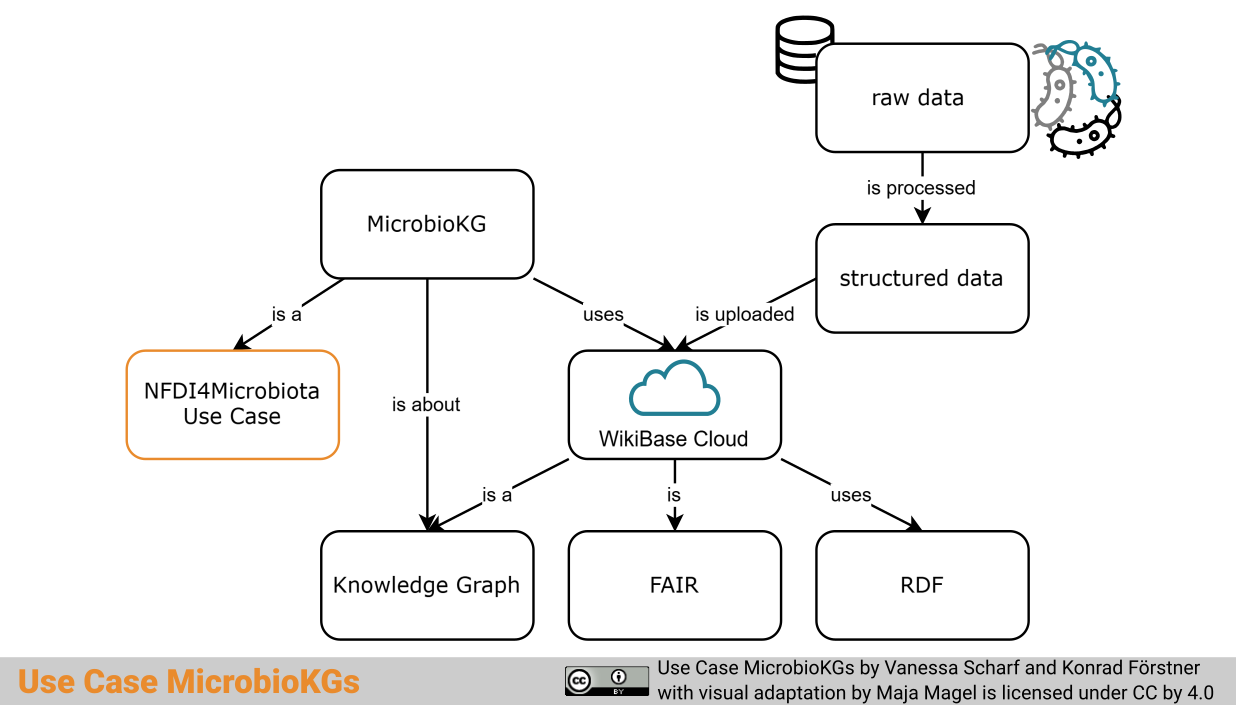
Short description
The representation of microbiological data in the form of an open Knowledge Graph provides search possibilities with the query language SPARQL and can enable the enrichment of data by linking it easily to other resources. After agreeing on a schema to fit the nature of your data, MicrobioKGs
serves as a blueprint on how to store your information in a findable, accessible, interoperable, and reusable way.
The Use Case MicrobioKGs aims at the semantic representation of heterogeneous and partially neglected data/ metadata from microbiology to enable powerful search and query opportunities. These will be showcased on three specific examples:
(1) Bacterial sRNAs and their regulatory network in different species,
(2) Metadata of numerous aquatic microbiome samples,
(3) Information of outbreaks of Q fever, an infectious disease caused by the bacterial pathogen Coxiella burnetii
Graphical abstract
Graphical abstract “Use Case MicrobioKGs” by Vanessa Scharf and Konrad Förstner.
How you can contribute
You are a …
- microbiologist who generates or works with microbiological data, and would like to make it accessible following the FAIR principles:
Learn to create items, define properties, and connect your data through semantic relationships
Planned output
Research output
- Working towards connecting microbiological data with domain-specific, yet currently not linked data
- Analyze linked data and gain potential insights by identifiying new correlations and relationships, e.g., by combining datasets with geolocation and sample types to track outbreak routes
Infrastructure output
- Using Wikibase, which offers an easy-to-use web interface as well as an API, enables research communities to maintain and curate their data
- Creating knowledge bases for future analysis in a cost-efficient manner
- Pilot knowledge graphs for different domains in microbiology
Training
- Demonstrate how users without a strong computational background can use Wikibase for curating their own datasets in a knowledge graph
Project Lead
Vanessa Scharf
ORCID ID: 0000-0002-8059-3664
ZB MED

Konrad Förstner
ORCID ID: 0000-0002-1481-2996
ZB MED
Keywords
knowledge graph
microbiology
metadata
domain specific graph databases
linked open data